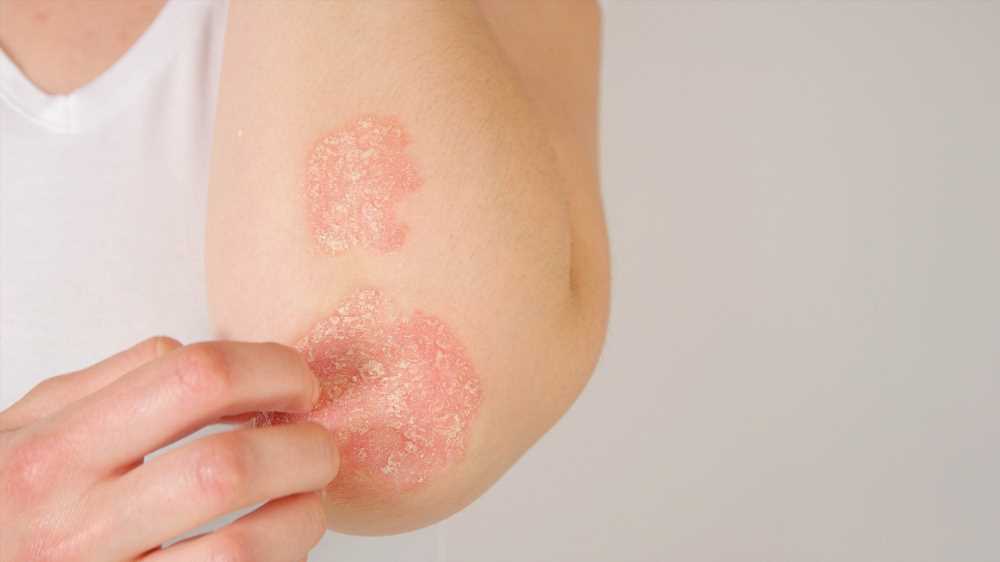

In a recent article posted to the Research Square preprint* server, researchers investigated the potential relationship between gut microbiota and different subtypes of psoriasis, including arthropathic psoriasis and psoriasis vulgaris.
The study used a bidirectional two-sample Mendelian randomization (MR) approach for their investigations.
 Study: Association between gut microbiota and psoriasis: a bidirectional two-sample Mendelian randomization study. Image Credit: Flystock/Shutterstock.com
Study: Association between gut microbiota and psoriasis: a bidirectional two-sample Mendelian randomization study. Image Credit: Flystock/Shutterstock.com

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
Per the World Health Organization (WHO) data, although psoriasis incidence varies globally, it is common in the general population worldwide, with current estimates suggesting that it has gripped 2–4% of the global population.
Psoriasis is a chronic inflammatory disease mainly characterized by skin lesions, and its other clinical symptoms include erythematous and pruritic plaques.
Given its complex etiology, psoriasis can also lead to inflammation in multiple organ systems, including cardiovascular, musculoskeletal, and gastrointestinal systems. An individual's genetics, smoking status, obesity, medication use, and past infections are the key factors influencing the development of psoriasis and its subsequent progression.
One of its subtypes, psoriatic arthritis, can even lead to severe disability, exacerbate psoriasis-related complications, and even cause death. Despite active research, the etiology of psoriasis and its subtypes remains elusive.
Moreover, currently, there is no treatment for psoriasis, and its therapy only suppresses the disease progression, severely limiting a patient's quality of life.
Research has suggested that gut microbiome metabolism affects the skin and exerts immune modulatory effects. The loss of beneficial bacteria and diversity of gut microbiome affects the skin through the gut-skin axis, which might have causal effects on psoriasis.
Scientific evidence showed that excessive activation of T helper (Th)1 and Th17 cells and a decrease in T regulatory (Treg) cell numbers disrupts host homeostasis leading to an increase in harmful bacteria, disruption to the functioning of the intestinal mucosal barrier, gut inflammation, and bacterial translocation.
Furthermore, research has shown an association between bacterial species, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, and Tenerictes Bacteroides with psoriasis onset.
The gut microbiome characteristics also varied among patients of psoriasis, psoriatic arthritis, and psoriasis vulgaris. The results were inconsistent across studies; thus, researchers could not assess whether there was a causal relationship between gut microbiota and psoriasis.
Thus, there is an unmet need for a better understanding of the gut microbiome status of psoriasis patients and its causal relationship with psoriasis. This data could help develop effective curative treatments for psoriasis.
About the study
In the present study, researchers used a recently acquired, large, multi-ethnic dataset of human gut microbiomes from the MiBioGen consortium, which conducted genome-wide association studies (GAWS) to investigate the relationship between host genetics and the gut microbiome.
It had 18,340 multi-national participants, but the highest representation was from European groups. Although the MiBioGen consortium had identified 211 bacterial taxa, they used a dataset of only 196 bacteria for the current study analysis.
The researchers obtained the dataset for psoriasis and its two subtypes from the FinnGen Consortium's summary statistical data or L1 dataset, which exclusively had populations representing European groups. It comprised thousands of cases and controls for each psoriatic condition.
The study contained 1,637 cases, 212,242 controls, with16,380,462 single nucleotide polymorphisms (SNPs) for arthropathic psoriasis.
Likewise, this dataset had 4,510 cases, 212,242 controls, with 16,380,464 SNPs for psoriasis, and 2,802 cases, 212,242 controls, with 16,380,459 SNPs for psoriasis vulgaris.
The team analyzed the aggregated statistical data using an MR approach to explore the potential causal relationship between the gut microbiome and psoriasis. SNPs with a threshold P-value of 1 x 10−5 worked as genetic instrumental variables in these MR analyses.
The researchers assessed instrument validity using the F-statistic, where a >10 threshold value helped avoid weak instrument bias.
Furthermore, the researchers used several MR-related statistical methods. Of those, inverse variance weighting (IVW) assumed all SNPs were valid instruments and provided the most precise causal estimates.
The MR-Egger method estimated causal effects via the Egger regression slope coefficient and addressed potential biases. Similarly, the Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) method identified horizontal pleiotropy and minimized its impact.
The weighted median and weighted model methods worked even when some SNPs were considered invalid. Finally, the team conducted leave-one-out analyses to ensure unbiased causal estimations.
Results
The MR analysis revealed that 18, 35, and 61 SNPs were causally associated with psoriasis, psoriasis vulgaris, and arthropathic psoriasis, respectively, with all having F-statistics >10, indicating no weak bias.
The authors noted that a higher genetically predicted abundance of Odoribactor protected against psoriasis, while that of Ruminiclostridium5 increased the risk of psoriasis, with odds ratios (OR) of 0.71 and 1.31, respectively, indicating moderate effect sizes.
Similarly, an increased genetically predicted relative abundance of Verrucomicrobiae class, Verrucomicrobiaceae family, and Akkermansia genus was causally associated with a higher risk of arthropathic psoriasis.
Furthermore, an increased genetically predicted relative abundance of Eubacterium fissicatena and Lactococcus bacterial groups and Actinomycetales taxa was causally associated with a higher risk of psoriasis vulgaris.
Thus, supplementation with probiotics or fecal microbiota transplantation could help relieve psoriasis symptoms by improving gut permeability and correcting gut dysbiosis. These interventions have shown promise against the recurrence of psoriasis in many clinical studies.
Conclusions
The MR approach allowed the researchers to fetch reliable causative effects of gut microbiota on psoriasis and its subtypes. It minimized the impact of external factors and avoided confounding or reverse causality.
Since the team leveraged data from a large GWAS meta-analysis, the dataset of this study had high statistical strength.
Thus, this study's findings provided novel and valuable insights into psoriasis prevention, management, and treatment using specific gut microbes (bacterial groups).
Microbial-based studies could provide more insights into psoriasis pathogenesis, offering novel mitigation and therapeutic strategies. More work is needed to uncover the exact mechanisms causally relating to gut microbiota and psoriasis.

 *Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: Research Square publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Preliminary scientific report.
Zhang, J. et al. (2023) "Association between gut microbiota and psoriasis: a bidirectional two-sample Mendelian randomization study". Research Square. doi: 10.21203/rs.3.rs-3251626/v1. https://www.researchsquare.com/article/rs-3251626/v1
Posted in: Medical Science News | Medical Research News | Medical Condition News
Tags: Arthritis, Bacteria, Cell, Chronic, Disability, Dysbiosis, Fecal Microbiota Transplantation, Genetic, Genetics, Genome, Inflammation, Inflammatory Disease, Metabolism, Microbiome, Musculoskeletal, Nucleotide, Obesity, Probiotics, Psoriasis, Psoriatic, Psoriatic Arthritis, Research, Single Nucleotide Polymorphisms, Skin, Smoking

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.