In a recent study posted to the Research Square* preprint server, which is under consideration at Scientific Reports, a team of researchers developed a novel human Alveolar Epithelial Lentivirus immortalized (hAELVi) cell infection model to estimate the replication potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and other respiratory viruses in the lower respiratory tract of humans.
Study: SARS-CoV-2 Omicron variant is attenuated for replication in a polarized human lung epithelial cell model. Image Credit: Naeblys / Shutterstock
Autopsy studies have shown that type I and type II pneumocytes in the lung, covering the vast majority of the lung surface, are the building blocks of the alveolar epithelium. Also, these cells express SARS-CoV-2 host factors, including the receptor angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine subtype 2 (TMPRSS2).
In previous studies, primary alveolar cells cultured at the air-liquid interface (ALI) have mimicked many facets of the alveolar epithelium partially; however, they quickly lose their in vivo phenotype and do not permit SARS-CoV-2 infection. Similarly, there are issues with several currently used other lung-derived cell lines, including A549, NCI-H1299, Calu-3 cell lines, as they model the alveolar epithelium imperfectly. Even lung organoids and explanted human lung tissue replicate wild-type SARS-CoV-2 only at low levels.
More importantly, since its emergence, cell models that might facilitate the assessment of the potential of Omicron to propagate in the human lower respiratory tract and thereby induce severe disease are urgently warranted.
About the study
In the present study, researchers studied severe SARS-CoV-2 infections in the lower human respiratory tract using immortalized hAELVi cell line. They derived immortalized hAELVi cell lines from the type I pneumocytes and cultured them at the ALI.
They used filter supports to facilitate differentiation of hAELVi cells under ALI conditions to a stratified, polarized epithelial cell layer with high barrier functions and more closely resembling human body cells in their gene expression patterns.
The hAELVi cells have been used in several prior research studies; however, according to the authors, no reports suggest their usage in virus research.
Study findings
The hAELVi cells were seeded and grown on permeable filter inserts under liquid-liquid conditions for 72 hours, following which the cell cultures were incubated for up to 28 days under ALI. Cells developed under ALI conditions attained a cubic shape; in many regions, a multilayered organization was seen at day 0 that pseudostratified or stratified to columnar epithelium at day 21.
Under ALI, these cells developed transepithelial electrical resistance (TEER) after 10 days, with a high maximum value of 6000 Ω*cm2 approximately 21 days after seeding. Immunoblot and enzyme-linked immunosorbent assay (ELISA) analyses showed that hAELVi cells upregulated expression of the ACE2 and the TMPRSS2 host receptors in 28 days when grown under ALI conditions.
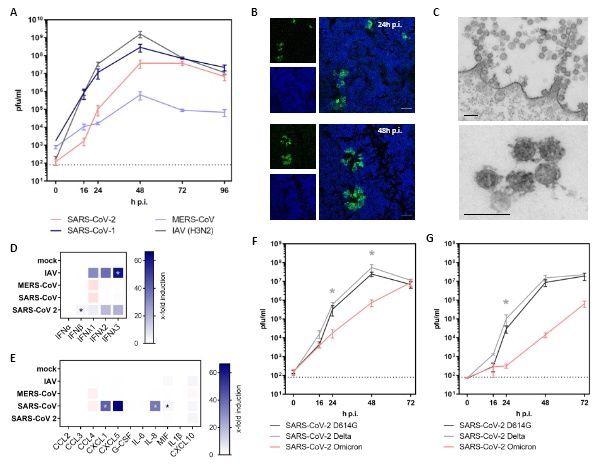
Infection of hAELVi cell air-liquid-interface cultures with highly pathogenic coronaviruses. (A-G) hAELVi cells grown under ALI for 21 days were used for infection experiments with highly pathogenic coronaviruses. (A) Cells were infected with SARS-CoV-2 D614G, SARS-CoV, MERS-CoV and IAV at MOI of 0.3 and further incubated under ALI conditions at 37°C. Progeny viruses were collected by washes form the apical compartment at indicated time points and titrated using standard plaque assay on VeroE6 cells. Replication analysis was performed for n=3 in technical duplicates. (B) Cells were infected with SARS-CoV-2 at MOI 1 and were processed for analysis by confocal laser-scanning fluorescence microscopy to detect the viral spike protein at 24h and 48h p.i. (green channel). Nuclei were stained with DAPI (blue channel). Data are representative for two independent experiments. Bar: 50μM (C) Thin-section electron microscopy of a SARS-CoV-2 infected epithelial cell shows cluster of coronavirus particles at the cell surface (upper image) and a small cluster at higher magnification. Infection was performed with MOI of 3 and cells were fixed 24h p.i.. Data are representative for two independent experiments. Bar: 200nM (D+E) Cells were infected with SARS-CoV-2, SARS-CoV, MERS-CoV and IAV at MOI of 1 followed by the collection of the basolateral fluid at 48h p.i. for ELISA detection of type I and III IFN (D) or the indicated cyto- and chemokines (E). Experiments were performed for three independent experiments in technical duplicates. (F+G) For growth curve analyses, cells were infected with MOI of 0.3 (F) or MOI 0.03 (G) with SARS-CoV-2 variants D614G, Delta or Omicron, respectively. Apical washes were performed at indicated time points and titrated using a standard plaque assay. Replication analysis was performed for n=2 in duplicates. Statistical analysis was done by using Kruskal-Wallis test, * p<0.05.
Growth curve analyses of progeny viruses harvested from apical washes of the ALI cultures revealed that the polarized hAELVi cultures were highly permissive to the D614 strain of SARS-CoV-2 with viral titers peaking at 48h post-infection (p.i.) at 5x 107 pfu/ml. Likewise, these cell cultures were permissive to seasonal influenza A virus (IAV), SARS-CoV, and Middle East respiratory syndrome (MERS-CoV).
Confocal laser scanning microscopy confirmed the infection by SARS-CoV-2 in mostly separated cells at 24h p.i. At 48 h p.i., infected cells formed larger clusters indicating ongoing viral cell-to-cell spread. Interestingly, the infected cell cultures showed all the hallmarks of SARS-CoV-2 replication, such as double-membrane vesicles, budding into membrane compartments, and release at the cell surface. ELISA results also showed that SARS-CoV-2 infection triggered modest secretion of interferon beta (IFN-β) and IF λ1-3, and immune factors, such as macrophage migration inhibitory factor (MIF) and C-X-C motif chemokine ligand 10 (CXCL10).
The analysis showed an attenuated growth phenotype of Omicron, reduced by up to two orders of magnitude at 48h p.i., compared to the Delta variant in the polarized human hAELVi lung cell cultures. At 48 p.i., this effect was even more pronounced, following infection with a 10-fold decreased multiplicity of infection (MOI) with the Omicron variant replicating to 1078-times lower titers than Delta.
Conclusions
Overall, the study highlighted that hAELVi cultures could be a good fit for fast phenotypic evaluation of emerging SARS-CoV-2 VOCs to inform public health strategies on time. Further, the findings evidenced that cultures of polarized hAELVI cells grown under ALI can model productive SARS-CoV-2 infections in the lower respiratory tract under controlled experimental conditions.
In the future, if the hAELVi cells could be co-cultured with macrophages-like cells, they present an opportunity for investigations of the cross-talk between epithelial and myeloid cells. Furthermore, SARS-CoV-2-infected hAELVi cultures could be used in studies examining the role of type I and type III IFN (antiviral cytokines) in tissue damage that has been observed in severely ill and critical coronavirus disease 2019 (COVID-19) patients.
*Important Notice
Preprints with Research Square publish preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Christin Mache, Jessica Schulze, Gudrun Holland, et al. SARS-CoV-2 Omicron variant is attenuated for replication in a polarized human lung epithelial cell model, 21 February 2022, PREPRINT (Version 1) available at Research Square https://doi.org/10.21203/rs.3.rs-1343204/v1, https://www.researchsquare.com/article/rs-1343204/v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Assay, Cell, Cell Line, Chemokine, Chemokines, Confocal laser scanning microscopy, Coronavirus, Coronavirus Disease COVID-19, covid-19, CXCL10, Cytokines, Electron, Electron Microscopy, Enzyme, Fluorescence, Fluorescence Microscopy, Gene, Gene Expression, in vivo, Influenza, Interferon, Lentivirus, Ligand, Lung Organoids, Macrophage, Magnification, Membrane, MERS-CoV, Microscopy, Omicron, Organoids, Phenotype, Protein, Public Health, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Serine, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Virus

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.
Source: Read Full Article