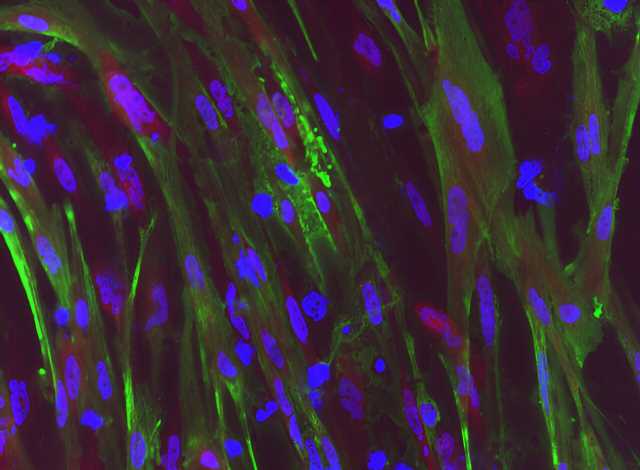
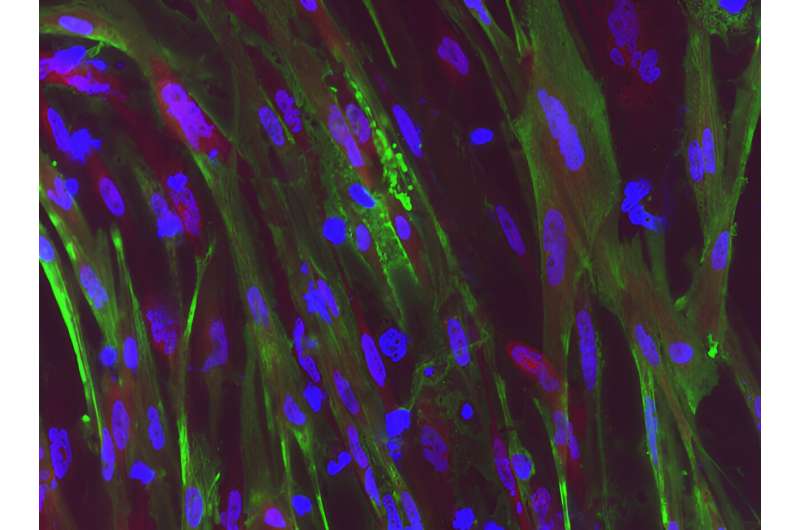
Duchenne muscular dystrophy (DMD) is a muscle degeneration disorder caused by mutations affecting the dystrophin gene. On August 24 in the journal Stem Cell Reports, researchers show how a dual CRISPR RNA method restored dystrophin protein function in induced pluripotent stem cells derived from DMD patients.
The approach worked by removing large sections of the dystrophin gene, allowing the cells to skip faulty or misaligned sections of the genetic code. This yields truncated but still functional proteins for a wide variety of mutation patterns associated with DMD.
“Dual CRISPR-Cas3 is a promising tool to induce a gigantic genomic deletion and restore dystrophin protein via multi-exon skipping induction,” says senior author Akitsu Hotta of Kyoto University. “We expect this study to enlighten new ways to treat DMD patients and other genetic disorders that require extensive deletions.”
Due to significant variations in the mutation patterns affecting the dystrophin gene, deleting a small section of the gene can only be used for a limited number of DMD patients. For example, the most common mono-exon skipping of exons 51, 53, and 45 can be applied to 13%, 8%, and 8% of DMD patients, respectively.
Multi-exon skipping (MES) has broad applicability to various DMD mutation patterns. By targeting the mutation hotspots in the dystrophin gene, MES from exon 45 to 55 was estimated to benefit more than 60% of DMD patients. Unfortunately, few techniques are available to induce a large deletion to cover the target exons spread over several hundred kilobases.
To overcome this hurdle, Hotta and his team used CRISPR-Cas3 to induce a deletion of up to 340 kilobases at the dystrophin exon 45-55 region in various DMD mutation patterns. Because it was rare to observe a deletion of more than a hundred kilobases using a single CRISPR RNA—which helps to locate the correct segment of DNA—the researchers used a pair of CRISPR RNAs inwardly sandwiching the target genomic region.
The authors note potential limitations of the dual CRISPR RNA system. First, there is variation in the deletion pattern, and the precise start and end points of the deletion cannot be fully controlled. This could be a drawback when a large but precise deletion is required. Second, the study did not demonstrate the functionality of the recovered dystrophin protein. Third, other methods should be developed to improve the overall genome editing efficiency of the Cas3 system.
“Our dual-Cas3 system might apply to future gene therapies once we’re able to deliver the dual-Cas3 components in vivo to skeletal muscle tissues safely and efficiently,” says Hotta. “The ability to induce several hundred kilobases of DNA deletion itself also has broad applicability for basic research when a large deletion is needed.”
More information:
Akitsu Hotta, Dual CRISPR-Cas3 system for inducing multi-exon skipping in DMD patient-derived iPSCs, Stem Cell Reports (2023). DOI: 10.1016/j.stemcr.2023.07.007. www.cell.com/stem-cell-reports … 2213-6711(23)00295-3
Journal information:
Stem Cell Reports
Source: Read Full Article