
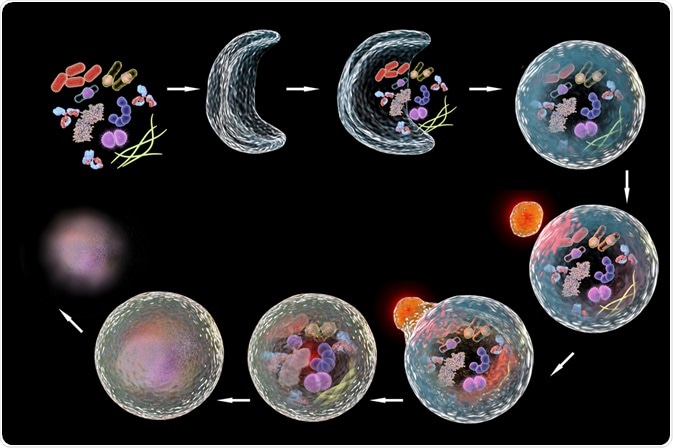
Autophagy is the process of regulated self-eating that occurs through the degradation of cellular components. This process is mediated by the sequestration of organelles or cytosolic fluid in vesicles which are bound by double membranes, called autophagosomes, and their subsequent fusion with degradative organelles called lysosomes or vacuoles.
 Kateryna Kon | Shutterstock
Kateryna Kon | Shutterstock
Lysosomes contain hydrolytic enzymes which cause the degradation of the contents transported by the autophagosome. The resultant macronutrients are recycled for use in the cytosol during starvation.
Yoshinori Ohsumi achieved a major breakthrough in the understanding of autophagy from a series of experiments on this process in yeast. This led to the discovery of the genes that regulate autophagy for which he was subsequently awarded a Nobel Prize in 2016.
Molecular machinery of autophagy
The process of autophagy is divided into distinct phases (1) initiation, (2) membrane nucleation and phagophore formation, (3) phagophore expansion, (4) fusion with the lysosome, and (5) degradation. Autophagy-Related (Atg) Proteins are responsible for the control and coordination of these phases and comprise the core machinery of the autophagy process.
The ATG proteins mediate the formation of the autophagosome and its delivery to the lysosome. There are five complexes composed of ATG proteins, and these are grouped by their functional and physical interactions:
- The ULK1 (Unc-51-like kinase 1) complex — comprised of the serine/threonine protein kinase ULK1, RB1-inducible coiled-coil protein 1 (FIP200), ATG13 and ATG101
- ATG9 — A single integral, transmembrane core ATG protein.
- The class III PI3K (PI3KC3) complex — consisting of class III PI3K, vacuolar protein sorting 34 (VPS34), Beclin 1, ATG14, activating molecule in Beclin 1-regulated autophagy protein 1 (AMBRA1) and general vesicular transport factor (p115)
- WIPI (WD repeat domain phosphoinositide-interacting) proteins and their interacting partner zinc-finger FYVE domain-containing protein 1 (DFCP1)
- ATG12 conjugated to ATG5, which in turn forms a complex with ATG16L and the ATG8s. The ATG8s include the light chain 3 (LC3) subfamily and the γ-aminobutyric acid receptor-associated protein (GABARAP) subfamily.
The key signals that initiate the autophagy cascade are those that indicate stress, such as hypoxia, protein aggregation and starvation. In starvation situations, the most notable protein mediators are mTOR and AMP-activated kinase (AMPK), both of which are highly conserved nutrient sensors.
These kinases, together with terminal kinases of various other stress-induced signaling pathways, converge on a common target, Unc-51-like kinase 1 (ULK1; Atg1 in yeasts) which consists of ULK1, autophagy-related protein 13 (ATG13), RB1-inducible coiled-coil protein 1 (FIP200) and ATG101.
Initiation of autophagy
The best-characterized trigger of autophagy induction is amino acid deprivation. Under such conditions, the master cell growth regulator serine/threonine kinase mTOR is inhibited. mTOR is typically found as a constituent of two complexes, mTORC1 and mTORC2, but only mTORC1 is involved with autophagy.
Under conditions of high nutrient availability, ATG13 and ULK1 are bound and phosphorylated by mTORC1 which results in their inactivation. However, during starvation, mTORC1 phosphorylated sites on ULK1 are dephosphorylated, and ULK1 dissociates. Concomitantly, ULK1 autophosphorylation occurs, followed by phosphorylation of ATG13 and FIP200 by ULK1.
It is important to note that there are several other regulators of the autophagy proteins other than mTOR, which is only described here by way of illustration.
Membrane nucleation and phagophore formation
Following autophagy induction, an organelle called the phagophore proceeds to nucleation. This occurs at a specific subcellular location termed the phagophore assembly site (PAS) on the ER. This is located on the endoplasmic reticulum (ER), as well as other organelles such as the Golgi complex, plasma membrane and recycling endosomes.
Nucleation of the phagophore membrane is complex. While the molecular mechanism has not been fully elucidated, it is known that phagophore formation involves PAS formation, together with ULK1 complex and PI3KC3–C1. This comprises of class III PI3K, vacuolar protein sorting 34 (VPS34), Beclin 1, ATG14, activating molecule in Beclin 1-regulated autophagy protein 1 (AMBRA1) and general vesicular transport factor (p115).
The PI3KC3–C1 must first be activated by one of its constituents, Beclin 1, which is inhibited by the antiapoptotic molecule BCL-2. The release of Beclin 1 by Bcl-2, which is triggered under stressful conditions, allows the class III PI3K (PI3KC3) complex to form. This complex phosphorylates a lipid called phosphoinositol, (PI) present in a characteristic ER structure called the omegasome. The resulting compound is phosphatidylinositol-3-phosphate (PI3P).
PI3P then recruits WIPIs and DFCP1 to the omegasome through their PI3P-binding domain. WIPI12 specifically binds to ATG16L1 directly, recruiting the ATG12~ATG5–ATG16L1 complex to the omegasome. This complex enhances the process of ATG3-mediated conjugation of ATG8s (which include the proteins LC3 and GABARAPs) to another lipid in the membrane called phosphatidylethanolamine (PE).
In this process, LC3 is cleaved by the protease ATG4 to form LC3-I, which is then conjugated with PE to form LC3-II. This conjugated form of LC3 (or GABARAPs) cause further recruitment of proteins that harbour an LC3-interacting region (LIR).
LC3/GABARAP interacts with proteins that facilitate the recruitment of cargo to the phagophore via autophagy receptors. The most well-understood autophagy receptors are p62, which recognizes proteins that have been tagged by the small protein ubiquitin.
Another notable autophagy receptor is BCL-2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3), which is a receptor for mitochondria via the process of mitophagy; the degradation of mitochondria by autophagy. The receptors are critical determiners of selectivity; hence this specific clearance of cargo, including organelles and macromolecules, is referred to as selective autophagy.
Phagophore expansion and sequestration of cytoplasmic cargo
ATG9 participates in the addition of membrane during elongation. Membranes to allow expansion of the phagosome are delivered by ATG9-containing vesicles. The autophagosomal membrane subsequently seals, producing a double-layered vesicle called the autophagosome, which undergoes maturation. This includes discarding the ATG proteins.
During maturation, machinery that mediates fusion with the lysosome, called SNARES, is recruited. SNARE proteins on the vesicle (v-SNAREs) and on the membrane of the lysosome target (t-SNAREs) combine to form a trans-SNARE complex that provides the force for membrane fusion.
ATG8s are another driver of autophagosome maturation, by linking the autophagosome to proteins called kinesins via an adaptor molecule. Kinesins are proteins that operate as motors in the cell, pulling their cargo along microtubules to their target location.
Fusion of the autophagosome
The fusion of the autophagosome with a lysosome produces an autolysosome. The SNARE proteins, as well as a protein called UVRAG, mediate this process. After the fusion event, degradation of sequestered cargo in the autolysosome takes place. This is achieved by the acidic hydrolases; the salvaged nutrients are released into the cytoplasm to be used again by the cell.
Triggers of autophagy
Autophagy is a highly selective and adaptive process that occurs in response to stress. Types of stress that are known to initiate the process include nutrient deprivation, growth factor depletion, hypoxia (low oxygen concentrations) and infection.
Initial understanding of the function of autophagy was as a means to provide nutrients to the cell during periods of scarcity induced by stressful conditions. As such, it was largely considered to be nonselective; more recent research has shown that it operates in a highly selective manner in the cell to target specific cytosolic material, such as damaged mitochondria or aggregated proteins, for selective autophagy. This additional function is called cytoprotection.
Sources
- https://www.nature.com/articles/s41580-018-0003-4#Sec1
- https://www.nature.com/articles/s41580-018-0033-y#Sec1
- www.sigmaaldrich.com/…/autophagy-signaling-pathways.pdf
- www.cellsignal.com/…/science-pathways-autophagy
- www.cellsignal.co.uk/…/pathways-autophagy
Further Reading
- All Autophagy Content
- What is Autophagy?
- Autophagy Process
- Autophagy Functions
- Autophagy Regulation
Last Updated: Feb 26, 2019

Written by
Hidaya Aliouche
Hidaya is a science communications enthusiast who has recently graduated and is embarking on a career in the science and medical copywriting. She has a B.Sc. in Biochemistry from The University of Manchester. She is passionate about writing and is particularly interested in microbiology, immunology, and biochemistry.
Source: Read Full Article