
An international team of 144 scientists studying lung function has made a huge leap in understanding chronic obstructive pulmonary disease by conducting the largest multi-ancestry, genome-wide association meta-analysis to date. The study, which included 580,869 participants, identified 1,020 independent disease association signals and implicated 559 genes from a variant-to-gene mapping framework. Most notably, these genes were enriched in 29 pathways, providing new insights into the underlying mechanisms of lung function.
In the study, “Multi-ancestry genome-wide association analyses improve resolution of genes and pathways influencing lung function and chronic obstructive pulmonary disease risk,” published in Nature Genetics, researchers investigated cell type and lung function in a genome-wide association study. Signals were sought between lung function and age, smoking and height.
Past studies have linked lung-function impairment to predictions of increased mortality from all causes. According to the World Health Organization, chronic obstructive pulmonary disease (COPD, emphysema or chronic bronchitis) is the third leading cause of death worldwide, causing 3.23 million deaths in 2019. While there is no cure for COPD, treatments and mitigation strategies (avoiding pollution and quitting smoking) can help.
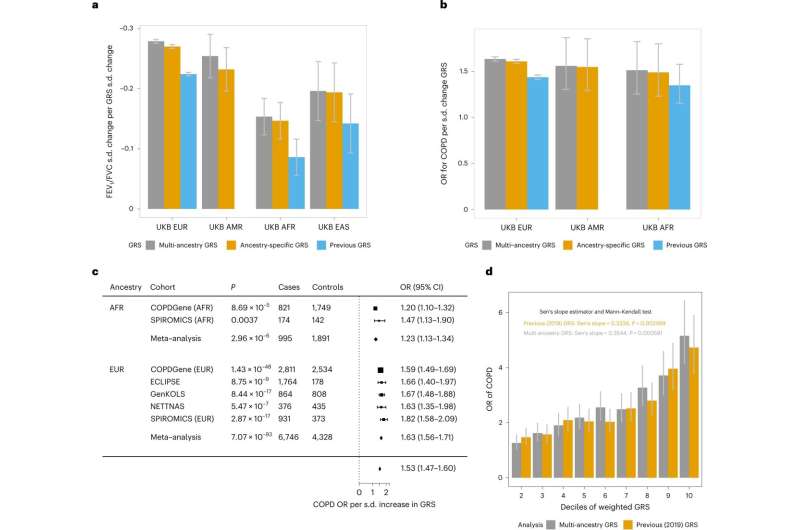
Abnormal tests are a criterion for diagnosing COPD. Lung function tests measure the amount of air the lungs can hold and how forcefully the air can be exhaled. The research team used lung function test data to run a meta-analysis across 66.8 million variants. The experiment identified 1,020 distinct signals for lung function, including 713 not described in previous studies. The signals mapped to 559 genes, highlighting pathways not previously implicated in lung function, suggesting they may play a critical role in lung health.
Aside from finding new genetic associations with COPD, the study also significantly improved understanding of past studies. Six previously implicated genes were supported by 14 additional criteria not seen in the original reports. Height, a determinant of lung growth, had no correlations between lung function associated signals. There were only nominal smoking-dependent effects on 69 signals, and 113 signals showed small evidence of age-dependent effects.
Any study involving humans is only as good as its sample size. Humans have such diverse lifestyles, diets, exercise routines, education histories, genetic predispositions and environmental pressures that a study attempting to say anything about all people (with a specific condition) needs to have a large number of participants. A study that finds a correlation between owning a cat and getting into car accidents in 58 people is indistinguishable from the noise of chance. In contrast, a study with more than 580,000 would be strong evidence that a signal has been found. Even with the robust sample, it would only be able to associate cat ownership with car accidents, but confidently enough to be the basis for future studies to examine how the observations are connected.
Additionally, the demographics of study participants are crucial. A study that finds no benefit from taking vitamin D supplements in winter may not seem definitive if it is conducted only on Sudanese farmers. Alternatively, an alarming report on the high mortality risks of running a marathon, conducted on lifelong smokers with a median age of 82, may have skewed contributing factors that do not apply to most people interested in running marathons.
In the current study, the sample size is well over half a million participants, which is exceptionally robust. Most of the participants (468,062) are of European ancestry, mainly because that is where most data is currently available. The multi-ancestry aspect comprised individuals of African (8,590), American/Hispanic (14,668), East Asian (85,279), and South Asian (4,270) ancestry. Ages ranged from children to adults across a diverse demographic, so the findings should be widely applicable.
The research paper concludes, “These findings bring us closer to understanding the mechanisms underlying lung function and chronic obstructive pulmonary disease and should inform functional genomics experiments and potentially future chronic obstructive pulmonary disease therapies.”
More information:
Nick Shrine et al, Multi-ancestry genome-wide association analyses improve resolution of genes and pathways influencing lung function and chronic obstructive pulmonary disease risk, Nature Genetics (2023). DOI: 10.1038/s41588-023-01314-0
Journal information:
Nature Genetics
Source: Read Full Article