
There is light at the end of the COVID-19 pandemic tunnel. Several vaccines against the SARS-CoV-2 virus that causes COVID-19 are now in clinical trials, with one—developed by Pfizer/BioNTech—already having been approved for emergency use in the United States. This has been the fastest development and rollout of any vaccine in history, starting with the first gene sequence released in January. (The previous record was held by the mumps vaccine, which took four years.)
But while this may seem like an overnight success story, the speed and effectiveness of these new vaccines can be in part attributed to the decades of research into infectious diseases that preceded the COVID-19 outbreak. Case in point: five of the vaccines, including those developed by Pfizer/BioNTech and Moderna, contain genetic mutations that increase their effectiveness, mutations based upon work dating back more than 10 years using the resources of the Advanced Photon Source (APS), a U.S. Department of Energy (DOE) Office of Science User Facility at DOE’s Argonne National Laboratory.
The APS has been at the forefront of the field of structural biology since its ultrabright X-ray beams first started shining in 1995. Scientists use those powerful beams of light to illuminate the structures of viruses, producing detailed pictures of the makeup of their proteins. Those pictures are then used to design techniques, drugs and vaccines that fight the virus more effectively.
Which is exactly what these mutations do. They were developed by Jason McLellan, now an associate professor at the University of Texas at Austin, and Barney Graham, currently the deputy director of the Vaccine Research Center at the National Institute of Allergy and Infectious Diseases, part of the National Institutes of Health (NIH). The pair’s current work on COVID-19 vaccines was heavily influenced by their research into an entirely different disease: respiratory syncytial virus (RSV), which affects thousands per year.
“We started that work in 2009,” said McLellan, who was then a postdoctoral researcher in Peter Kwong’s lab at NIH. “It took a while, but we had success developing a vaccine candidate for RSV in 2013.”
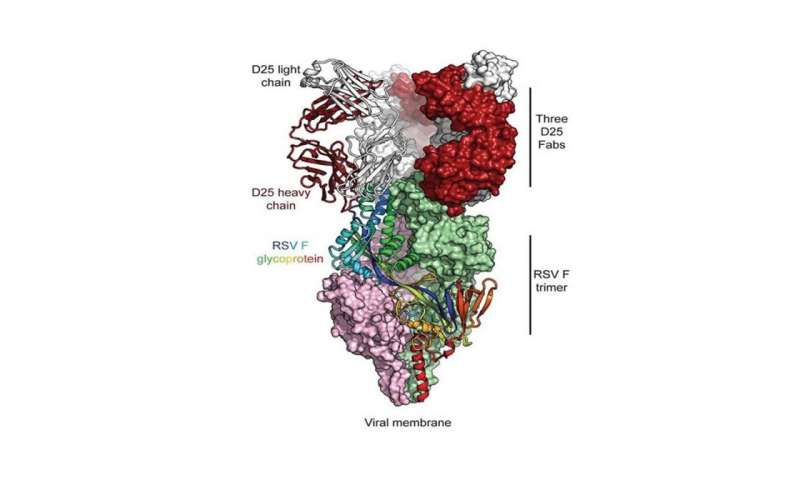
The key to that vaccine, which is now entering phase 3 trials, was a better understanding of one of the virus’s proteins. The F protein in RSV adorns the surface of the virus and makes first contact with human cells, infecting them. This protein has two shapes: prefusion, a smaller mushroom-like form that first makes contact with cells, and postfusion, an extended form that initiates entry of the virus into those cells. Once the protein is in its postfusion shape, infection has begun, and it is more difficult for the immune system to fight the virus off.
McLellan and Graham worked on the theory that a vaccine directed at the F protein in its prefusion mode should be more effective. They created more than 100 different variants of that protein to find a version that they could stabilize in the prefusion form. This would provide a target for the immune system, helping it to develop neutralizing antibodies against the virus. Parts of that work were done at the Southeast Regional Collaborative Access Team (SER-CAT) beamline at the APS, operated by the University of Georgia.
In May 2013, the pair and their colleagues reported success, publishing their work in Science. In November 2013, they reported a vaccine candidate for RSV that neutralized the F protein, stabilizing it in its prefusion form. That paper was also published in Science, and included structural work performed at SER-CAT. In 2019 McLellan, Graham and their colleagues published the clinical proof-of-concept in Science that showed the prefusion molecule was a better vaccine antigen.
McLellan and Graham soon learned that the technique they discovered could be applied to coronaviruses.
“Barney and I started thinking about what to look at next, and coronaviruses in general were similar to RSV,” McLellan said. “Both contain a similar protein: the spike in coronaviruses is like the F protein in RSV, except that the spike is about two and a half times the size. It’s like the F protein on steroids.”
In 2013, McLellan and his colleagues, including Nianshuang Wang, now at Regeneron Pharmaceuticals, began working with Graham and his NIH colleague Kizzmekia Corbett on a vaccine for the Middle East Respiratory Syndrome coronavirus (MERS-CoV). The team applied the same technique to MERS-CoV, and in 2017 reported similar success. This time they performed some of their work at the Structural Biology Center (SBC) at the APS, and published results in the Proceedings of the National Academy of Sciences.
“Both McLellan and Graham are longtime users of the APS, and their work to understand RSV and MERS-CoV led to an effective technique to neutralize both viruses,” said Argonne’s Bob Fischetti, group leader and life sciences advisor to the APS director. “The structural work performed at the APS played a key role in the discovery of this technique, and we are now seeing how important it might be.”
When SARS-CoV-2 emerged, McLellan, Wang and graduate student Daniel Wrapp, now at the University of Texas at Austin, joined with Graham and Corbett to see if the same technique would work to inhibit this new virus’s spread. And once they were able to examine the structure of the spike protein, they discovered that it did.
“Being able to solve the structure of the SARS-CoV-2 spike protein and know that it was in the desired conformation within three weeks of sequence release was critical for rapidly advancing vaccine development and antibody discovery programs,” Graham said.
The initial results of their work, a mutation called S-2P, is now in SARS-CoV-2 vaccines developed by Moderna, Pfizer/BioNTech, Novavax, Johnson & Johnson and CureVac, McLellan confirmed. Pfizer’s and Moderna’s vaccines use messenger RNA (mRNA) to instruct human cells to make the mutant of the spike protein that the immune system then develops antibodies against. If a person is infected with the virus, the immune system then recognizes it before infection and the antibodies neutralize the spike protein, by binding to it and preventing it from infecting the cell.
The U.S. Food and Drug Administration granted an emergency use authorization for Pfizer’s vaccine on Dec. 12, and is expected to make their recommendation regarding Moderna’s vaccine this week. Pfizer’s vaccine is now being administered in the United States.
“The previous work allowed us to move very quickly,” McLellan said. “Vaccine development usually takes a decade. The RSV vaccine we created in 2013 is just now entering phase 3 clinical trials. COVID-19 went from a genome sequence to first immunization in less than a year. We played a role in the development of the antigen. There was a massive response to this coronavirus. Lots of labs and companies contributed.”
McLellan and his colleagues published further work on SARS-CoV-2 in Science in September. Again the scientists tested more than 100 variations of the spike protein, this time using a technique called cryogenic electron microscopy at their own labs. They ended up with a molecule called HexaPro that could be 10 times as potent as the S-2P spike protein in the current COVID-19 vaccines. It is currently being evaluated for use by several companies.
While the prospect of several COVID-19 vaccines is exciting, McLellan is similarly heartened by what this accelerated development process means for the future. We don’t know what the next pathogens are going to be, he notes, and the tools and techniques being developed now will play an important role.
“We have learned a lot about the different steps involved in a pandemic response,” he said. “The vaccine development platform will continue to mature. The speed and new technology saves us days and weeks, and helps with the game plan to respond to future pandemics.”
Graham and Corbett were recently named recipients of a Golden Goose Award for their work on COVID-19 research, and McLellan’s work is recognized in the announcement.
The APS has dedicated more than 10,000 hours of time to research into the structure of SARS-CoV-2 this year, and has made its resources available to more than 80 groups of researchers from across the country, in addition to the work done by Argonne scientists there. With more than 100 structures of the virus made available to the world scientific community—more than any other U.S. light source—the APS has been an important factor in the effort to combat COVID-19.
Source: Read Full Article