
Phage display is a molecular tool used to produce and analyze polypeptide function and strucure by presenting them on the surface of lysogenic filamentous bacteriophages.
 Design_Cells | Shutterstock
Design_Cells | Shutterstock
Lysogeny
The term lysogenic refers to the ability of bacteriophages to assume a dormant state, referred to as a prophage, in the bacterial cell. Lysogeny is characterised by the integration of the viral DNA into the host bacterium's genome.
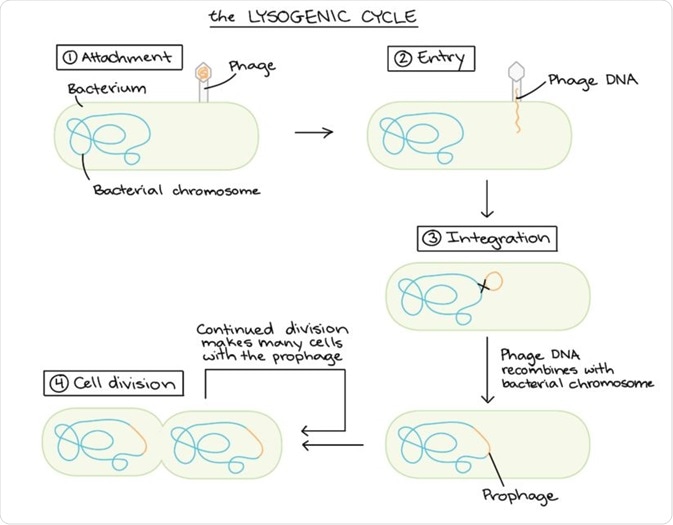
Schematic diagram of the lysogenic cycle. (Image modified from "Conjugation," by Adenosine (CC BY-SA 3.0). The modified image is licensed under a CC BY-SA 3.0 license).
Phage display
Phage display integrates the gene encoding the polypeptide to be displayed into the host DNA. Thus, the polypeptide is expressed as a fusion product with the phage coat protein.
This concept can be expanded to create peptide libraries of up to 1010 different fusion variants that can be used to study enzymology, protein-ligand interactions, determine the site for antibody and receptor binding or identify the epitopes for monoclonal antibodies, and to determine the identity of enzymes substrate.
Phage display systems
The most common vehicle for phage display are E.coli filamentous bacteriophages (f1, fd, M13). The three phages share 98.5% of their DNA sequence and are thus closely related.
The peptides and antibodies are displayed on the capsid which is composed of the major capsid protein (gp/pVIII), of which there are several thousand copies, and four minor capsid proteins (gp/pIII, VI, VII, and IX) present in pairs at the tips of the virus in single copy numbers. The variability in copy numbers and inherent properties of each capsid protein enables several configurations of the displayed protein/peptide.
Displays can be tethered on either the N- or C- at both termini of the capsid protein at various densities. Small and high density display proteins are suited to the major capsid protein pVIII as a display vehicle; display of larger proteins are suited for low density display.
A display protein on minor phage proteins is achievable for monovalent (binds one target) or polyvalent (binds many targets) proteins.
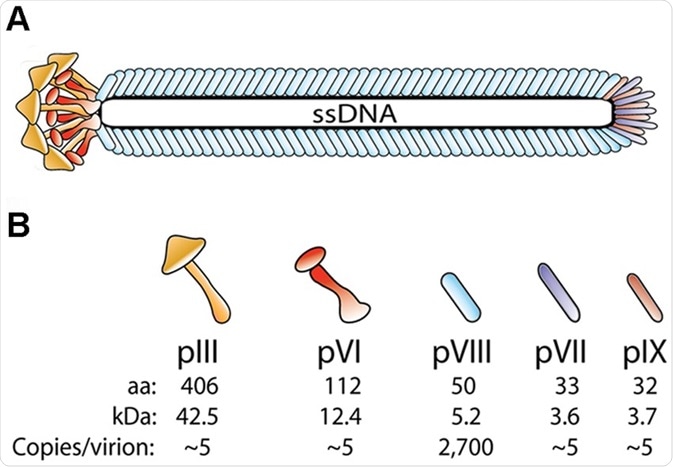
Schematic drawing showing bacteriophage structure. (A) The virion coat is comprised of 5 structural proteins that encircle the viral genome (ssDNA) (B) There are approximately 2,700 copies of pVIII and approximately 3–5 copies each of the four proteins pIII, pVI, pVII and pIX which are located in pairs, at the tips (N-termin) and ends (C-termini) of the capsid protein.
Phage display: key steps
Phage display is a multistep process beginning with the production of a gene library.
Gene library
The gene encodes the display protein of interest and is associated with multiple regulatory sequences which optimise the process of replication, transcription, translation, and presentation of the display protein.
Cloning
The display protein is cloned into a vector that possess characteristics of both bacteriophage and plasmids, called phagemid. It contains two origins of replication; a– bacterial origin of replication which allows the propagation of the plasmid in the E. coli and an F1 phage origin for replication and packaging of single-stranded DNA (ssDNA).
This allows the phagemid vector to be packaged into the capsid of a bacteriophage. A helper phage is necessary to assist the assembly of the mature virions with their corresponding protein fragments.
Screening
Screening is used to identify high affinity binding partners of the display protein. Phage that present the protein of interest are put in contact with the ligand of interest which is immobilised on a microtiter plate. This allows binding of the applied phages to the mobilised ligands.
A wash step then eliminates non-specific binding and the adsorbed (bound) phages are eluted. The phagemid is then transformed in E. coli cells for amplification. This allows the virus to undergo lysogeny and produce multiple copies of the virus with the display protein joined, or conjugated to the structural proteins of the coat. The process allows only the highest affinity ligands to bind.
Applications of phage display
Phage display is used to discover peptides and proteins in order to discover better mechanisms of drug discovery and delivery, production of diagnostic tests for disease monitoring, and evaluating the efficacy of treatments. The libraries of genetically modified phages are central to proteomic, immunological and cellular research.
The process of linking a gene encoding a protein of interest with the viral coat protein gene causes the phage to phenotypically display the protein on the coat; the encoding gene remains in the phage genome.
The process links genotype (genetic constituent that encodes a characteristic) and phenotype (observable characteristics), allowing identification of the coding sequences of binding proteins.
Sources:
- www.whatisbiotechnology.org/…/phage-display-helps-study-the-interaction-of-proteins
- https://www.sciencedirect.com/science/article/pii/S2221169117305130
- www.scielo.br/scielo.php
Further Reading
- All Bacteriophage Content
- What are Bacteriophages?
- Phage Display: Applications in Research
- Temperate Bacteriophages and the Lysogenic Cycle
Last Updated: Nov 13, 2018

Written by
Hidaya Aliouche
Hidaya is a science communications enthusiast who has recently graduated and is embarking on a career in the science and medical copywriting. She has a B.Sc. in Biochemistry from The University of Manchester. She is passionate about writing and is particularly interested in microbiology, immunology, and biochemistry.
Source: Read Full Article